Mathematical modeling of cell fate decisions to understand cancer biology

Mathematical models serve to explain complex biological phenomena and provide predictions that can be tested experimentally. They can provide plausible scenarios of a complex biological behaviour when intuition is not sufficient anymore.
Mathematical models can provide the appropriate tools to interpret all sorts of experimental observations in a rigorous and systematic manner.
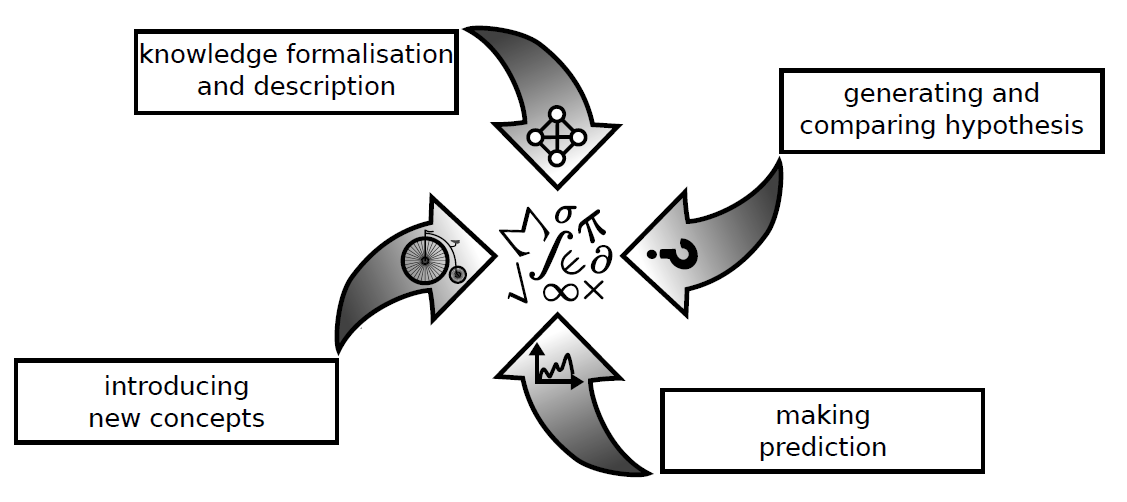
More specifically, we propose four characteristics or hallmarks of a mathematical model:
• A model facilitates the formalisation of our knowledge: it aims at describing biological phenomena in a formal and unambiguous way; it recapitulates (integrates all facts) and summarises (provides concise description) what is known about a biological process; it allows the identification and the listing of the key players and mechanisms involved in a process; it also provides a way to visualise biological data;
• A model helps the generation and comparison of hypotheses: it enables analyses and interpretation of biological data; this leads in turn to the formulation of hypotheses on the network structure, on the plausible molecular interactions behind a process;
• A model formulates predictions: it can predict results such as mutant phenotypes, response to drug treatments, identification of therapeutic intervention points, behaviours in particular cellular contexts; it can also test and rank predictions by performing in silico experiments; it can anticipate side-effects of drugs or perturbations;
• A model enables conceptualization: it can be used as an abstract thinking object and it allows introduction of new concepts in biology: in particular it can give formal definition for a class of behaviours and cellular observations (e.g., any dynamical behaviour of individual motifs)
Over the past years, we have developed logical models that focus on cell fate decisions, cell cycle, apoptosis, EMT and migration, etc. with the purpose of understanding which mechanisms are deregulated in some particular contexts and that can lead to cancer.
Highlights

Cohen DP, Martignetti L, Robine S, Barillot E, Zinovyev A, Calzone L. Mathematical Modelling of Molecular Pathways Enabling Tumour Cell Invasion and Migration. PLoS Comput Biol. 2015. 11(11):e1004571
November 3, 2015

Remy E, Rebouissou S, Chaouiya C, Zinovyev A, Radvanyi F, Calzone L. A Modeling Approach to Explain Mutually Exclusive and Co-Occurring Genetic Alterations in Bladder Tumorigenesis.Cancer Res. 2015. 75(19):4042-52.
October 1, 2015 | Mechanistical model of mutual exclusive genome alterations

Chanrion M, Kuperstein I, Barriè re C, El Marjou F, Cohen D, Vignjevic D, Stimmer L, Paul-Gilloteaux P, Biè che I, Tavares Sdos R, Boccia GF, Cacheux W, Meseure D, Fre S, Martignetti L, Legoix-Né P, Girard E, Fetler L, Barillot E, Louvard D, Zinovyev A, Robine S. Concomitant Notch activation and p53 deletion trigger epithelial-to-mesenchymal transition and metastasis in mouse gut. 2014. Nat Commun. 5:5005.
October 2014 | Experimentally validated model prediction

Kutumova E, Zinovyev A, Sharipov R, Kolpakov F.Model composition through model reduction: a combined model of CD95 and NF-kappaB signaling pathways. 2013. BMC Syst Biol. 7(1):13.
February 2013 | Composite model of apoptosis

Stoll G, Surdez D, Tirode F, Laud K, Barillot E, Zinovyev A, Delattre O. Systems biology of Ewing sarcoma: a network model of EWS-FLI1 effect on proliferation and apoptosis. 2013. Nucleic Acids Res., 41(19):8853-71.
July 2013 | Network model of Ewing sarcoma

Zinovyev A, Kuperstein I, Barillot E, Heyer W-D.Synthetic Lethality between Gene Defects Affecting a Single Non-essential Molecular Pathway with Reversible Steps.. 2013. PLoS Comput Biol 9(4): e1003016.
April 2013 | Model of within-pathway synthetic lethality

Barillot E., Calzone L., Hupe P., Vert J.-P., Zinovyev A. Computational Systems Biology of Cancer Chapman & Hall, CRC Mathematical & Computational Biology, 2012, 452 p.
August 2012 | Book written from the team experience

Morozova N., Zinovyev A., Nonne N., Pritchard L.-L., Gorban A.N., Harel-Bellan A. Kinetic signatures of microRNA modes of action 2012. RNA 18(9): 032284.
September 2012 | First team publication on mathematical modeling a biological journal

Calzone L, Tournier L, Fourquet S, Thieffry D, Zhivotovsky B, Barillot E, Zinovyev A. Mathematical modelling of cell-fate decision in response to death receptor engagement. 2010. PLoS Comput Biol 5;6(3):e1000702.
March 2010 | >100 citations

Gorban AN, Radulescu O, Zinovyev AY.Asymptotology of chemical reaction networks. 2010. Chemical Engineering Science 65, 2310-2324.
July 2010 | Principles of model reduction

Zinovyev A, Morozova N, Nonne N, Barillot E, Harel-Bellan A, Gorban AN.Dynamical modeling of microRNA action on the protein translation process. 2010. BMC Systems Biology 4:13.
February 2010 | Model of MiRNA mechanisms

Radulescu O., Gorban A., Zinovyev A., Lilienbaum A.Robust simplifications of multiscale biochemical networks. 2008. BMC Systems Biology 2:86.
October 2008 | Mathematical model of NfKb pathway